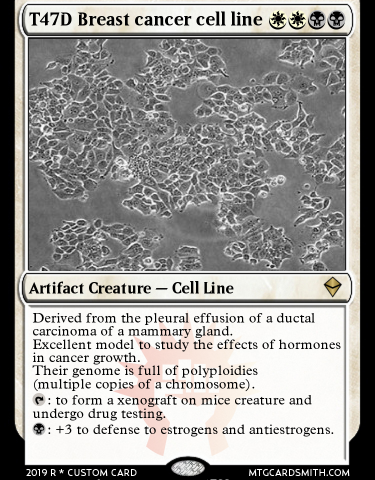
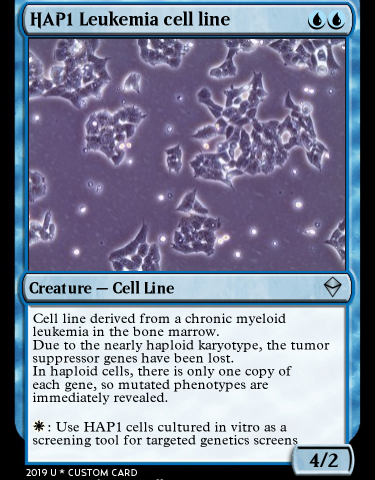
In order to select a set of two cancer cell lines on which we will perform the 3D genomics experiments, we conducted a survey among researchers at CRG. Those were the two most voted options and will be the first cell lines analyzed through Genigma.
Our priority will be T47D. This cell line is one of the most used in the 4D Genome Project, an ERC synergy project granted to four group leaders at the CRG. In the 4D Genome we study from different perspectives how the chromatin 3D conformation changes inside the cell nucleus during induced temporal transformations embryo development stages. For instance, François Le Dily in Miguel Beato’s lab treated T47D cells using steroid hormone action in T47D cells, they and explored how eukaryotic the genome of these cells reorganize in response to this particular external cues stimulus. Specifically, they study how signals are transduced to the nucleus and modulate chromatin structure and gene expression. (ref. 1).
We we also work with HAP1. The main advantage of this cell lines is that it is nearly haploid. This means that they have each cell nucleus contains a single copy of each of the chromosomes, and not two or more copies as usual of each chromosome. This feature makes HAP1 cells an excellent candidate for the first stages of the Genigma project, because of the it may be easier simplicity to get obtain their genome map from them. HAP1 is a cell line used in Eva Novoa’s new group at the CRG, where they are trying, among other goals, to characterize the role of RNA modifications in human disease, synaptic plasticity and transgenerational inheritance.
Lots of discoveries have been and will be, possible using cancer cell lines as models. However, a precise map of their genome is not still available for most of them and researchers have been using the map of a normal cell, which is not accurate 100%. To obtain an accurate map of cancer cells is our goal in Genigma. Are you in?
Credits:
Images retrieved and adpted from ATCC lgcstandards-atcc.org/ and Horizon horizondiscovery.com/ cell line repositories. Both web pages consulted on May 7th , 2019. Part of the text has been adapted from Wikipedia entries for T47D and HAP1. Cards created with https://mtgcardsmith.com/mtg-card-maker/
References: Le Dily, F et al. Hormone-Control Regions Mediate Steroid Receptor-Dependent Genome Organization. Genome Res 29 29-39. 01/01/2019.http://dx.doi.org/10.1101/gr.243824.118